Visual Med-Alpaca: Bridging Modalities in Biomedical Language Models
| Chang Shu1* | Baian Chen2* | Fangyu Liu1 | Zihao Fu1 | Ehsan Shareghi 3 | Nigel Collier1 |
| 1University of Cambridge 2Ruiping Health 3University of Monash |
Demo
Overview
Domain-specific foundation models are crucial in the biomedical field as the language used in biomedical text is highly specialized and contains numerous domain-specific terms and concepts not found in general domain text corpora like Wikipedia and Books. Pre-training on significant amounts of biomedical text has been shown to enhance the performance of language models on various biomedical text mining tasks when compared to existing publicly available biomedical PLMs. However, with the large number of parameters in modern language models, the cost of fine-tuning even a 7B model solely on PubMed is too expensive for most academic institutions that lack sufficient computing resources. Pre-training models on extensive medical image datasets to acquire multi-modal abilities is even more costly. Therefore, more cost-effective techniques such as Adapter, Instruct-Tuning and Prompt Augmentation are being explored to develop a model that can be trained and deployed on gaming-level graphics cards while still possessing sufficient capabilities. Furthermore, there is no public multimodal foundation model designed for biomedical usage to the best of our knowledge. As a result, we are pleased to release the Visual Med-Alpaca, an open-source, multi-modal, biomedical foundation model. Visual Med-Alpaca uses a prompt manager to merge the textual and visual information into the prompt for generating responses with biomedical expertise. The model is fine-tuned using two distinct datasets to incorporate biomedical knowledge and visual modality. The process involves collecting inquiries from various medical datasets and synthesizing answers with a gpt-3.5-turbo model. The study involves the integration of visual foundation models, namely the DEPLOT and Med-GIT models, to accommodate medical images as inputs. The Med-GIT model is a GIT fine-tuned specifically on the ROCO dataset to facilitate specialized radiology image captioning. The training procedure for the model is available in the Github repository. Visual input is a critical element of the medical domain, and the system architecture is designed to facilitate the seamless integration of alternate medical visual foundation models. The model's architecture is developed to translate image to text, followed by a cognitive engagement in reasoning over the text thereby derived. The most important task in the future is to systematically evaluate the medical proficiency and potential defects of Visual Med-Alpaca, including but not limited to misleading medical advice, incorrect medical information, etc. In addition to the conventional use of benchmarking and manual evaluation, we hope to target different model users (doctors and patients) and evaluate all aspects of the model in a user-centred manner. It is also important to note that Visual Med-Alpaca is strictly intended for academic research purposes and not legally approved for medical use in any country. Resources:
We apologize for the inconvenience, but this project is currently undergoing internal ethical screening at Cambridge University. We anticipate releasing the following assets within the next 1-2 weeks. You are more than welcome to Join Our Waitlist, and we'll notify you as soon as they become available.
Model Architecture and Training Pipeline
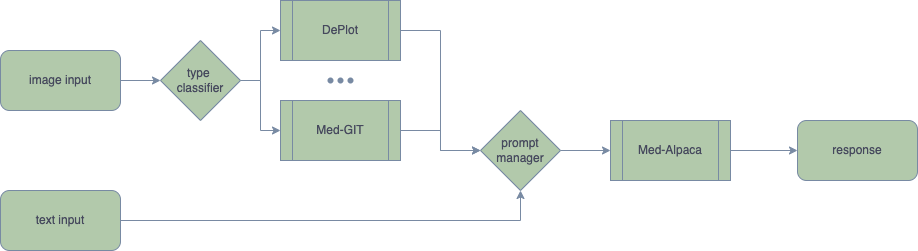
Domain Adaptation: Self-Instruct in Biomedical Domain
The process of collecting inquiries from various medical question-and-answer datasets (MEDIQA RQE, MedQA, MedDialog, MEDIQA QA, PubMedQA) is implemented in our study. This approach aims to increase the diversity and thoroughness of the dataset and improve the accuracy and comprehensiveness of the obtained results.
We synthesize answers of these questions with gpt-3.5-turbo in the self-instruct fashion. The gpt-3.5-turbo model is equipped with advanced natural language processing capabilities that enable it to understand and generate human-like responses to a wide range of questions. This makes it a reliable tool for generating structural and informative answers.
The process of filtering and editing question-answer pairs was performed manually. A total of 54,000 turns were carefully selected, taking into account the criteria of balance and diversity.
Visual Adaptation: Medical Image Captioning and Deplot
Visual input is a critical element of the medical domain, contributing essential information in healthcare settings. Healthcare practitioners heavily rely on visual cues to diagnose, monitor and treat patients. Medical imaging technologies, such as X-ray, CT and MRI, provide an unparalleled means of examining internal organs, identifying diseases and abnormalities that may not be visible to the naked eye.
Our study involves a further development of our previous work on visual language reasoning concerning charts and plots, as showcased in DEPLOT: One-shot visual language reasoning by plot-to-table translation. In this study, we enhance our approach by incorporating a visual foundation model that is capable of accommodating radiology images as inputs.
Within this particular framework, the task of visual language reasoning can be delineated into a bifurcation consisiting of two key phases: (1) the process of translating image to text, followed by (2) a cognitive engagement in reasoning over the text thereby derived.
The process involves the utilization of visual foundation models to convert medical images into an intermediate text state. The converted data is subsequently employed to prompt a pre-trained large language model (LLM), relying on the few-shot reasoning abilities inherent in LLMs.
At present, our platform is capable of supporting two distinct visual foundation models, namely the DEPLOT and Med-GIT models, considering the prevalence of plot and radiology imagery within the medical field. This system's architecture is also designed to facilitate the seamless integration of alternate medical visual foundation models.
The Med-GIT model represents a GIT: Generative Image-to-text Transformer for Vision and Language, fine-tuned specifically on the ROCO dataset to facilitate specialized radiology image captioning. The training procedure for the model is outlined in comprehensive detail in our publicly accessible Github repository.
Case Study
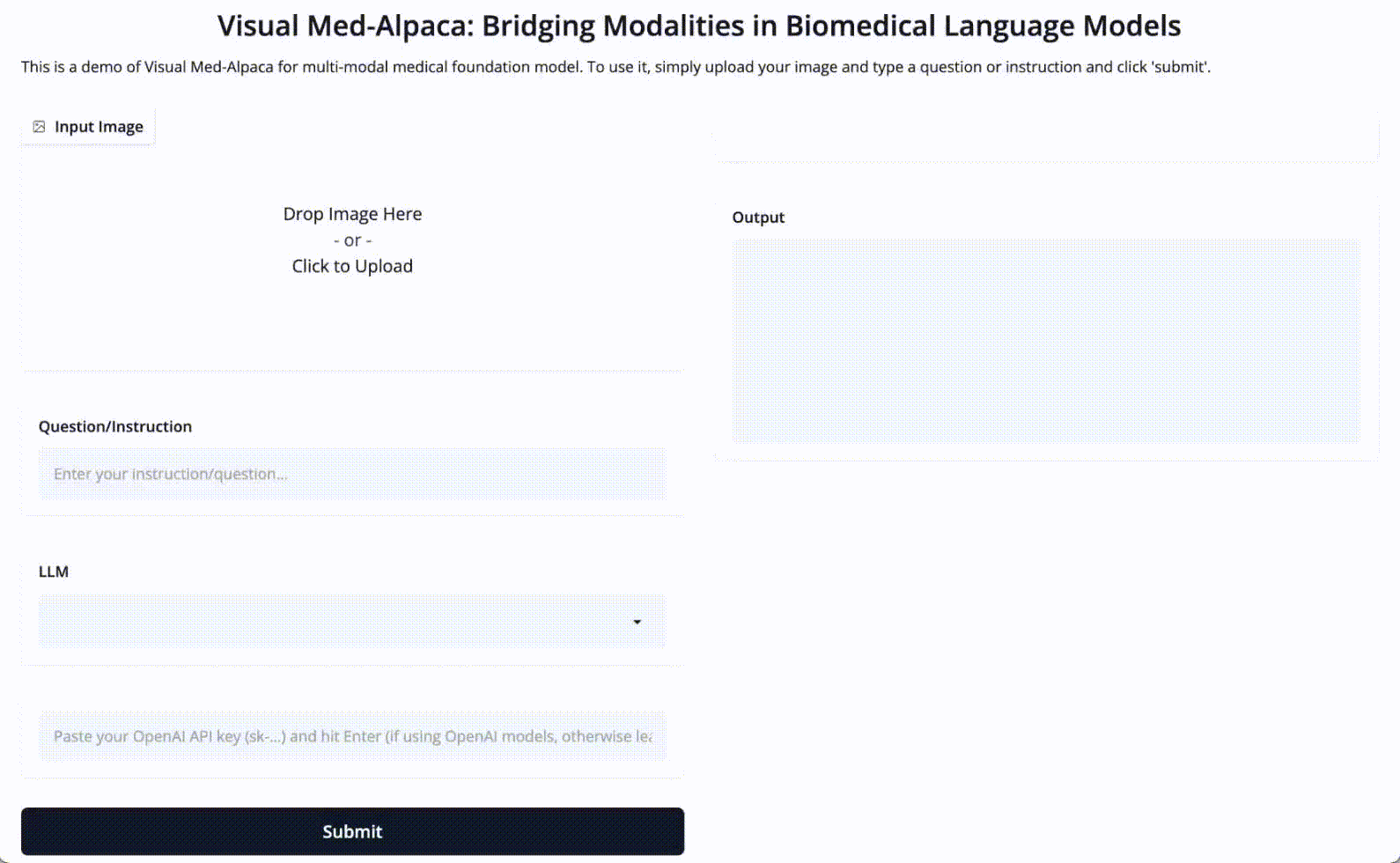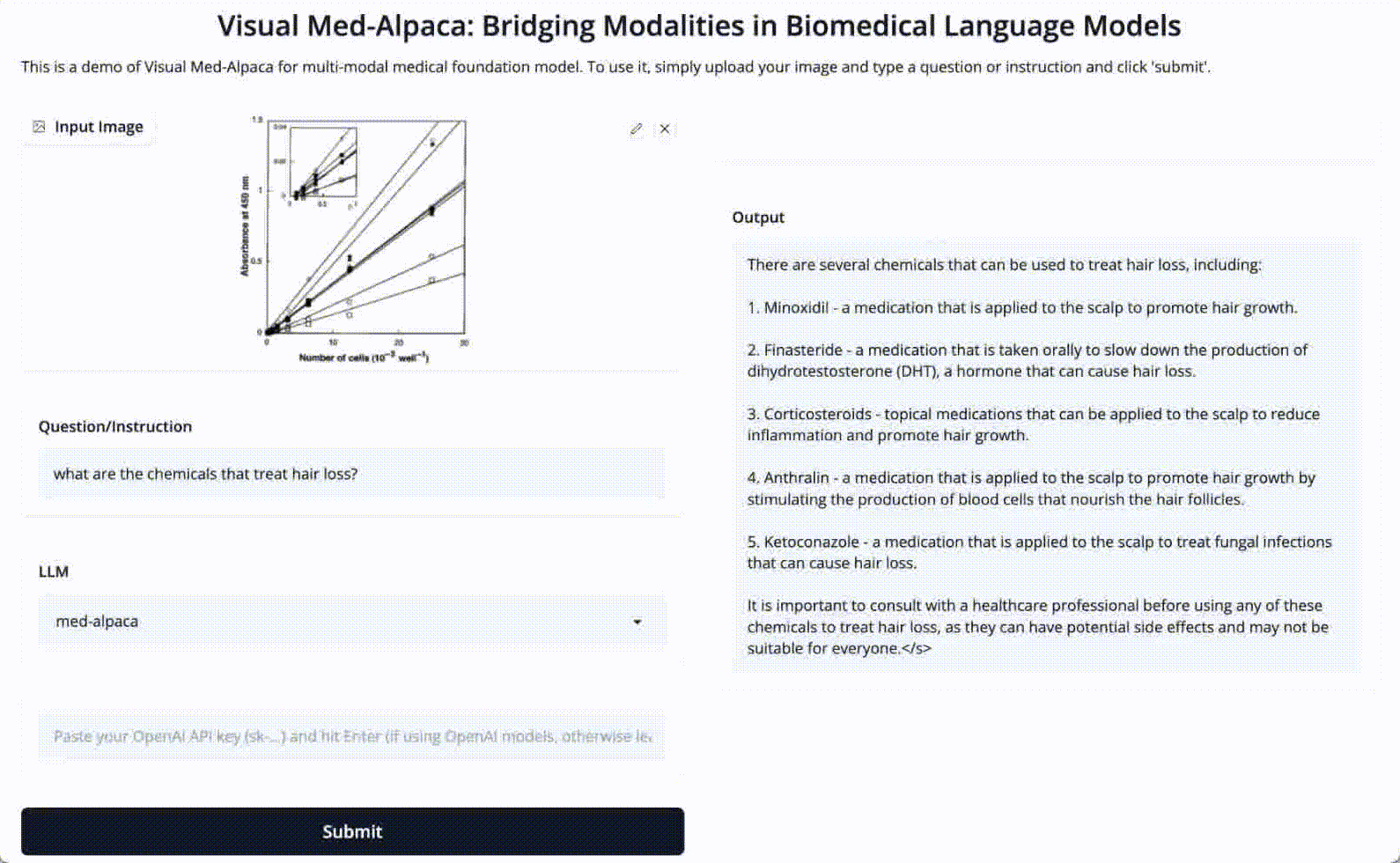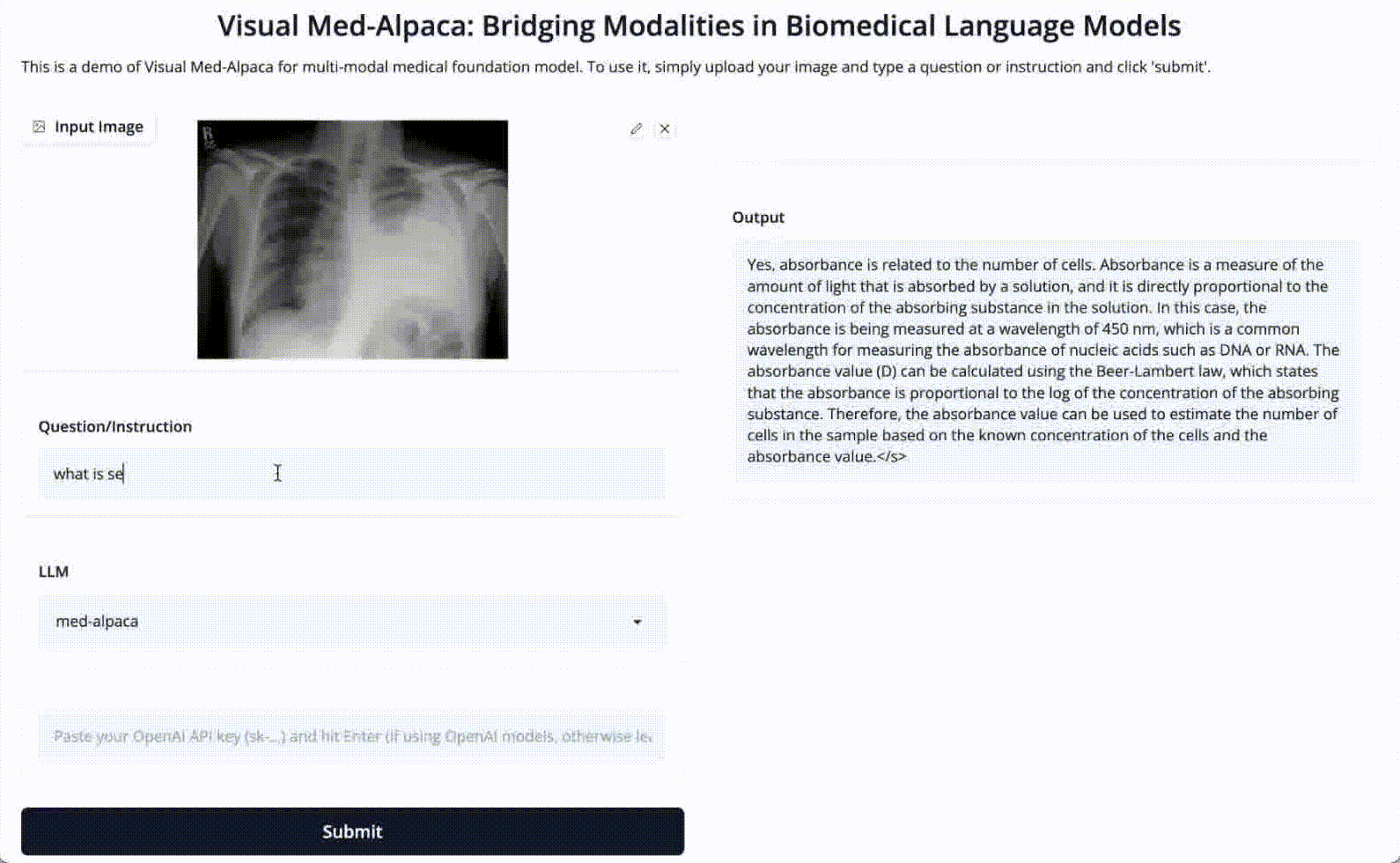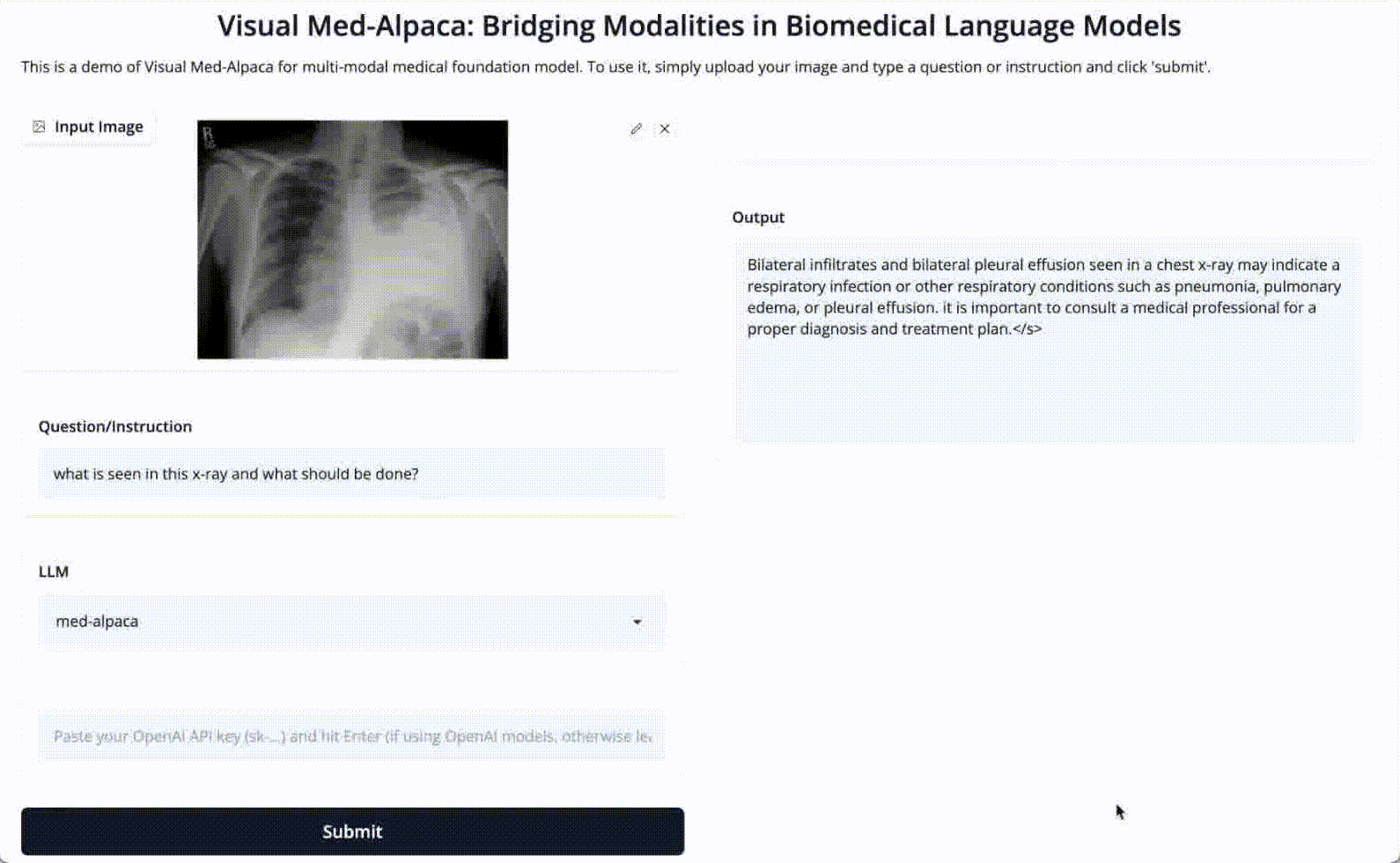
Input 1: What are the chemicals that treat hair loss?
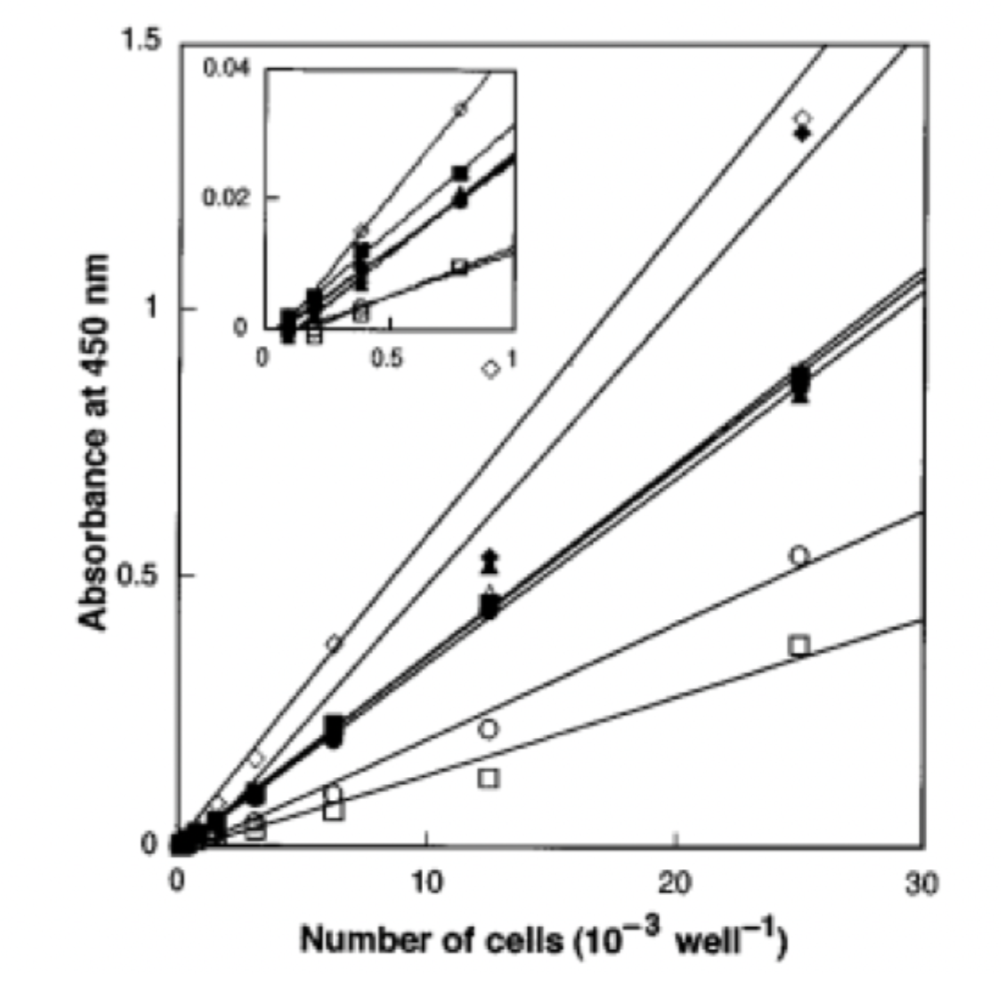
Input 2: Is absorbance related to number of cells?
Image:
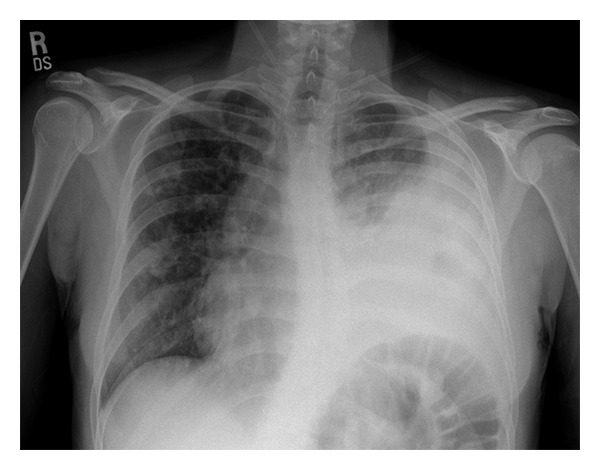
Input 3: What is seen in the X-ray and what should be done?
Image:
Future Work
One of the most crucial future works is the systematic evaluation of Visual Med-Alpaca, as well as other NLP models within the biomedical field. With the varying structure and type of medical data, it is essential to assess the efficacy of NLP models and their generalizability across different data sets.
We also expect pretraining on medical data can enhance the performance of NLP models in the biomedical field. It should help in the identification and reasoning of disease phenotypes, drug mechanism and the representation of clinical concepts.
The addition of genome protein modality may also help in achieving better reasoning in NLP models. Given that genetic and protein information are critical for understanding disease processes, NLP can aid in the analysis of large volumes of genomic data, making it possible to identify novel mutations involved in various disease processes. Therefore, incorporating genomic information into NLP models will enable a wider range of applications within the biomedical field.
Implementation Details
We follow the hyper-parameters as reported in the Github repo of Alpaca-LoRA and Alpaca:
Disclaimers
Visual Med-Alpaca, is intended for academic research purposes only. Any commercial or clinical use of the model is strictly prohibited. This decision is based on the License Agreement inherited from LLaMA, on which the model is built. Additionally, Visual Med-Alpaca is not legally approved for medical use in any country. Users should be aware of the model's limitations in terms of medical knowledge and the possibility of misinformation. Therefore, any reliance on Visual Med-Alpaca for medical decision-making is at the user's own risk.
Note: The developers and owners of the model, the Language Technology Lab at Cambridge University, do not assume any liability for the accuracy or completeness of the information provided by Visual Med-Alpaca, nor will they be responsible for any potential harm caused by the misuse of the model.
Acknowledgement
We are deeply grateful for the contributions made by open-source projects:
LLaMA,
Stanford Alpaca,
Alpaca-LoRA,
Deplot,
BigBio,
ROCO,
Visual-ChatGPT,
GenerativeImage2Text.
|